I am a computational biophysicist with a deep interest in understanding the biological processes that drive cellular life. My research focuses on developing and applying high-performance computational methods to uncover how complex biomolecular assemblies form, transform, and function. Through my postdoctoral work in the Department of Biophysics at Johns Hopkins University, I have created open-source software and parallelized algorithms for large-scale particle-based reaction-diffusion simulations. These tools enable modeling of dynamic protein assemblies, helping to study fundamental processes such as clathrin-mediated endocytosis and HIV virus lattice assembly. During my doctoral studies at the Chinese Academy of Sciences, I built theoretical frameworks and computational models to study the stepping mechanisms of diverse kinesin motors, providing insight into their distinct yet related functional behaviors. By integrating algorithms development, modeling and simulation, my research aims to bridge the gap between molecular level details and cellular level behavior.
📝 Publications
Protein Self-assembly
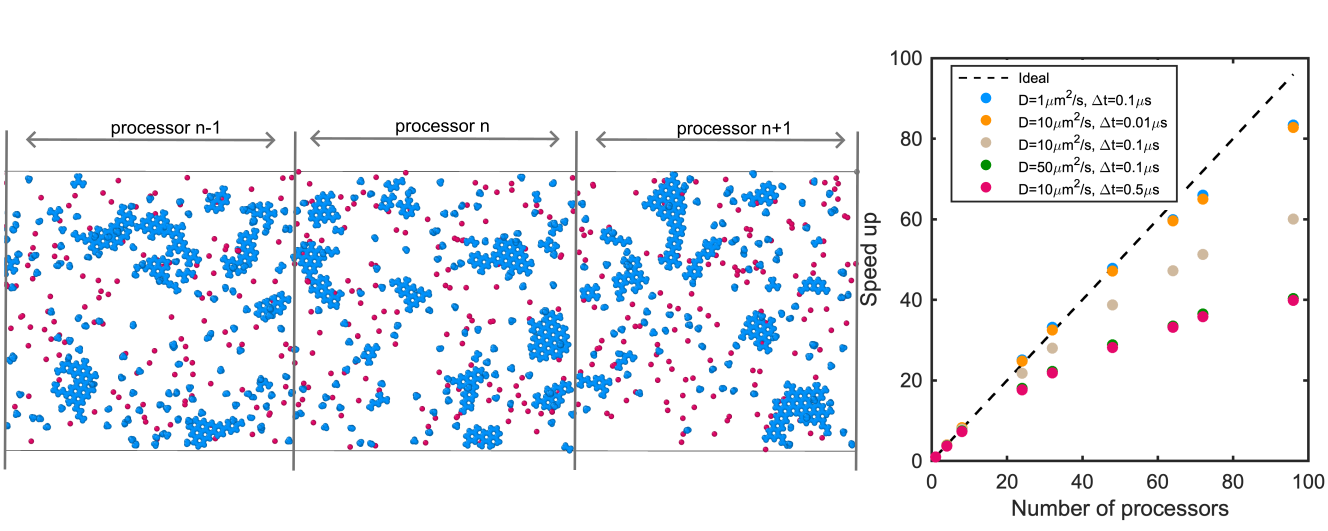
Parallelization of particle-based reaction-diffusion simulations using MPI
Sikao Guo, Korolija Nenad, Milfeld Kent, Jhaveri Adip, Sang Mankun, Ying M Yue, Margaret E Johnson, bioRxiv. 2024.12.06.627287 (2024).
By developing a parallel implementation of NERDSS using MPI-based domain decomposition, We have achieved near-linear scaling on large computing clusters. This enables the simulation of complex, biologically relevant self-assembly processes—such as protein lattice formation and the organization of multimeric complexes. Although performance depends on system size, reaction networks, and timescales, our open-source, parallelized code significantly improves the speed and accessibility of structure-resolved reaction-diffusion simulations, providing a powerful resource for researchers studying molecular assembly.
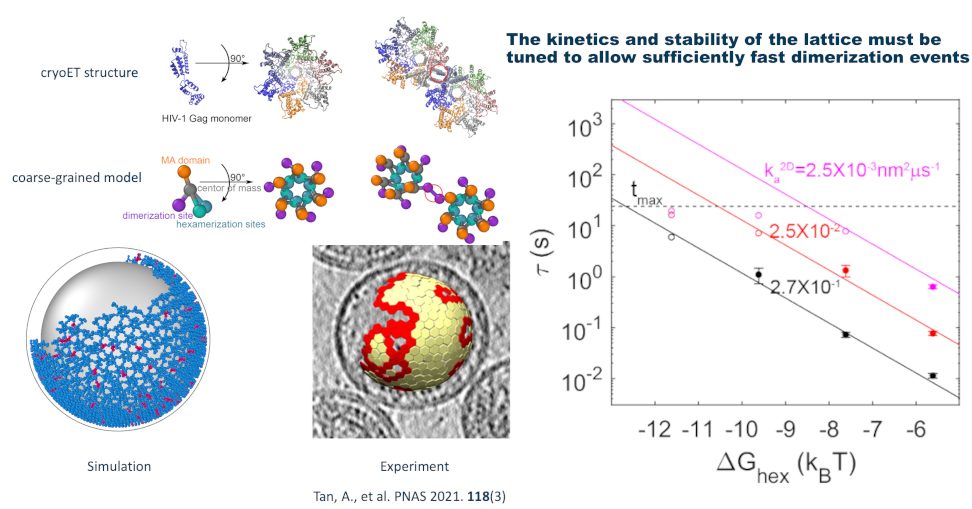
Structure of the HIV immature lattice allows for essential lattice remodeling within budded virions
Sikao Guo, Ipsita Saha, Saveez Saffarian, Margaret E Johnson, eLife. 12:e84881 (2023).
“This fundamental work substantially advances our understanding of the maturation of retroviruses, a key step in understanding the formation of infectious viruses. The evidence supporting the conclusions is compelling, with rigorous computational simulations. The work will be of broad interest to the community of virologists worldwide.” – Editor’s evaluation
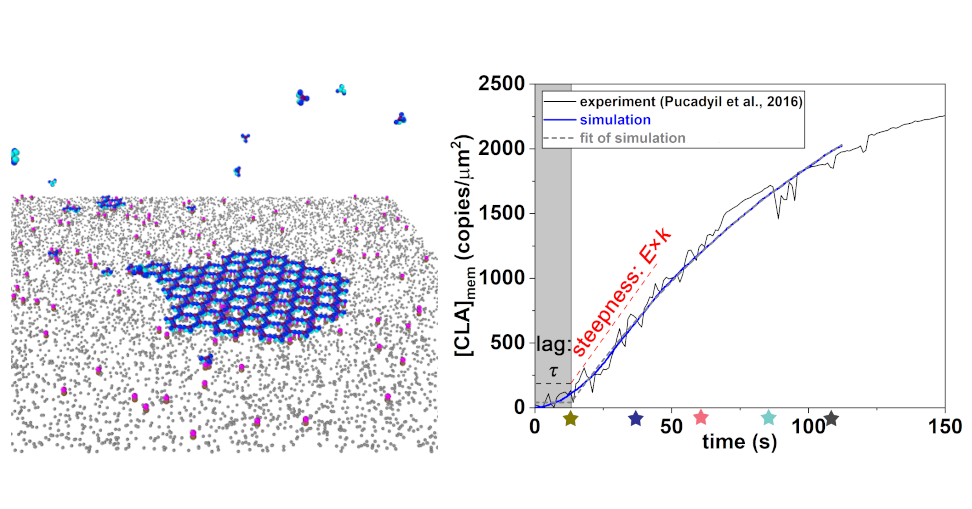
Large self-assembled clathrin lattices spontaneously disassemble without sufficient adaptor proteins
Si-Kao Guo, Alexander J Sodt, Margaret E Johnson, PLOS Computational Biology. 18, e1009969 (2022).
We explore how clathrin-coated structures assemble and disassemble on cell membranes, a key process in cellular transport. By integrating in vitro kinetic data with detailed reaction-diffusion simulations and membrane modeling, We show that stable nucleation of these lattices requires a greater-than-1:1 ratio of adaptor proteins to clathrin. Under more physiologically relevant conditions, insufficient adaptor abundance leads to spontaneously forming but ultimately transient assemblies, explaining why so many clathrin structures fail to mature into vesicles. This work provides a quantitative framework for understanding how cellular conditions govern the transition from transient clathrin-coated structures to productive vesicle formation.
-
Dynamin1 long- and short-tail isoforms exploit distinct recruitment and spatial patterns to form endocytic nanoclusters, 1. Jiang A., Kudo, K., Gormal R. S., Ellis S., Sikao Guo, Wallis T. P., Longfield S. F., Robinson P. J., Johnson M. E., Joensuu M., Meunier F. A., Nature Communications, 15: 4060, DOI: https://doi.org/10.1038/s41467-024-47677-8 (2024).
-
Temporal control by cofactors prevents kinetic trapping in retroviral Gag lattice assembly, Yian Qian, Daniel Evans, Bhavya Mishra, Yiben Fu, Zixiu Hugh Liu, Sikao Guo, Margaret E Johnson, Biophysical Journal DOI:https://doi.org/10.1016/j.bpj.2023.06.021 (2023).
-
Secretion-Catalyzed Assembly of Protein Biomaterials on a Bacterial Membrane Surface, Qi Xie, Sea On Lee, Nitya Vissamsetti, Sikao Guo, Margaret E. Johnson, Stephen D. Fried, Angewandte Chemie DOI:https://doi.org/10.1002/ange.202305178 (2023).
Molecular motor: Kinesin
- A common chemomechanical coupling model for orphan and conventional kinesin molecular motors, Si-Kao Guo, Ping Xie, Biophysical Chemistry 264, 106427 (2020).
- All‐atom molecular dynamics simulations reveal how kinesin transits from one‐head‐bound to two‐heads‐bound state, Xiao‐Xuan Shi, Si‐Kao Guo, Peng‐Ye Wang, Hong Chen, Ping Xie, Proteins: Structure, Function, and Bioinformatics 88 (4), 545-557 (2020).
- Run length distribution of dimerized kinesin-3 molecular motors: comparison with dimeric kinesin-1, Si-Kao Guo, Xiao-Xuan Shi, Peng-Ye Wang, Ping Xie, Scientific reports 9, 16973 (2019).
- Force dependence of unbinding rate of kinesin motor during its processive movement on microtubule, Si-Kao Guo, Xiao-Xuan Shi, Peng-Ye Wang, Ping Xie, Biophysical Chemistry 253, 106216 (2019).
- Dynamics of cooperative cargo transport by two elastically coupled kinesin motors, Yi-Ben Fu, Si-Kao Guo, Peng-Ye Wang, Ping Xie, The European Physical Journal E 42 (4), 1-13 (2019).
- A generalized kinetic model for coupling between stepping and ATP hydrolysis of kinesin molecular motors, Ping Xie, Si-Kao Guo, Hong Chen, International journal of molecular sciences 20 (19), 4911 (2019).
- Force dependence of velocity and run length of kinesin-1, kinesin-2 and kinesin-5 family molecular motors, Si-Kao Guo, Wei-Chi Wang, Peng-Ye Wang, Ping Xie, Molecules 24 (2), 287 (2019).
- ATP-concentration-and force-dependent chemomechanical coupling of kinesin molecular motors, Ping Xie, Si-Kao Guo, Hong Chen, Journal of Chemical Information and Modeling 59 (1), 360-372 (2018).
- Investigating role of conformational changes of microtubule in regulating its binding affinity to kinesin by all‐atom molecular dynamics simulation, Xiao‐Xuan Shi, Yi‐Ben Fu, Si‐Kao Guo, Peng‐Ye Wang, Hong Chen, Ping Xie, Proteins: Structure, Function, and Bioinformatics 86 (11), 1127-1139 (2018).
- Processivity of dimeric kinesin‐1 molecular motors, Si‐Kao Guo, Xiao‐Xuan Shi, Peng‐Ye Wang, Ping Xie, FEBS Open Bio 8 (8), 1332-1351 (2018).
- Dynamics of dimeric kinesins: Limping, effect of longitudinal force, effects of neck linker extension and mutation, and comparison between kinesin-1 and kinesin-2, Si-Kao Guo, Peng-Ye Wang, Ping Xie, International journal of biological macromolecules 105, 1126-1137 (2017).
- A model of processive movement of dimeric kinesin, Si-Kao Guo, Peng-Ye Wang, Ping Xie, Journal of Theoretical Biology 414, 62-75 (2017).
Professional Experience
01/2020 - present
Postdoctoral Research, Johns Hopkins University, Department of Biophysics
Research Advisor: Professor Margaret E. Johnson
Educations
09/2014 - 12/2019
Ph.D. in Condensed Matter Physics, Institute of Physics, Chinese Academy of Sciences, Beijing, China
Research Advisor: Professor Ping Xie
Graduate Thesis Title: “Studying the movement mechanism of dimer kinesin through theoretical modeling and numerical simulation”
09/2010 - 06/2014
B.S. in Physics, Nankai University, Tianjin, China
Conference Presentations
- Defects Within the HIV-1 Immature Lattice Support Dynamic Remodeling and Protease Dimerization, Cell Bio 2022, Washington, DC, USA, December 4, 2022.
- Large Self-assembled Clathrin Lattices Spontaneously Disassemble Without Sufficient Adaptor Proteins, Biophysical Society 66th Annual Meeting, San Francisco, CA, USA, February 19, 2022.
- Self-assembled clathrin lattices spontaneously disassemble without sufficient links to the plasma membrane, Stochastic Physics in Biology (Gordon Research Conference), Ventura, CA, USA, October 10, 2021.
- Modeling Nucleation and Kinetics of Clathrin Assembly by Membrane Localization, Biophysical Society 65th Annual Meeting, Virtual, February 22, 2021.
- Molecular mechanism of dimeric kinesin, 2018 Annual Summary Report, Institute of Physics, Chinese Academy of Sciences, Beijing, China, November 25, 2018.
- A model of processive movement of dimeric kinesin, Year-end Meeting and Exhibition of IOP, Institute of Physics, Chinese Academy of Science, Beijing, China, January 5, 2018.
Academic Service
- Reviewer of The Journal of Physical Chemistry, PROTEINS: Structure, Function, and Bioinformatics, PLOS Computational Biology, International Journal of Molecular Sciences, Cells, Applied Sciences, Biophysica, Biochemistry and Biophysics Reports, Entropy.
Open-Source Software
- Developer of a simulator for self-assembly at the cellular scale – NERDSS, Parallel NERDSS.
- Developer of the Python library for setting up models and analyzing output of the reaction-diffusion simulation Repo, PyPi
- Creator and developer of the simulation kit for molecular motor – kinesin, link1, link2, link3.
Contact
Department of Biophysics, Johns Hopkins University Baltimore, MD 21218
sikaoguo@gmail.com
